Deeper understanding of the myocardial structure linking the morphology and function of the heart would unravel crucial knowledge for medical and surgical clinical procedures and studies. Several conceptual models of myocardial fiber organization have been proposed but the lack of an automatic and objective methodology prevented an agreement. We sought to deepen this knowledge through advanced computer graphical representations of the myocardial fiber architecture by diffusion tensor magnetic resonance imaging.
MethodsWe performed automatic tractography reconstruction of unsegmented diffusion tensor magnetic resonance imaging datasets of canine heart from the public database of the Johns Hopkins University. Full-scale tractographies have been built with 200 seeds and are composed by streamlines computed on the vector field of primary eigenvectors at the diffusion tensor volumes. We also introduced a novel multiscale visualization technique in order to obtain a simplified tractography. This methodology retains the main geometric features of the fiber tracts, making it easier to decipher the main properties of the architectural organization of the heart.
ResultsOutput analysis of our tractographic representations showed exact correlation with low-level details of myocardial architecture, but also with the more abstract conceptualization of a continuous helical ventricular myocardial fiber array.
ConclusionsObjective analysis of myocardial architecture by an automated method, including the entire myocardium and using several 3-dimensional levels of complexity, reveals a continuous helical myocardial fiber arrangement of both right and left ventricles, supporting the anatomical model of the helical ventricular myocardial band described by F. Torrent-Guasp.
Keywords
It is widely accepted that the myocardial fiber architecture plays a critical role in many functional aspects of the heart, such as electrical propagation1,2 or ventricular contraction.3,4 It is also accepted that myocardium, as well as its fibers, may undergo architectural alterations in many heart diseases,5,6 leading to inefficient heart function. However, there is a lack of consensus about the exact distribution of the myocardial fibers and their spatial arrangement that constitutes the gross (left and right ventricles) myocardial structure. Deeper understanding of the precise cardiac architecture7 and its relationship to ventricular function8 would benefit clinical procedures such as surgery planning in left ventricular reconstructive surgery or resynchronization therapies.9,10
Researchers have proposed at least 7 conceptual models11 in attempts to accurately describe the architecture of the heart from dissection or histological procedures. Two of the most controversial approaches are the cardiac mesh model proposed by Anderson et al.12,13 and the helical ventricular myocardial band (HVMB) proposed by Torrent-Guasp,14 and Torrent-Guasp et al.15 The cardiac mesh model proposes that the myocytes are arranged longitudinally and radially, changing angulations along with myocardial depth and binding this architectural disposition to a functional one.16 On the other side, the HVMB model states that the ventricular myocardium is a continuous anatomical helical layout of myocardial fibers, linking the ventricular anatomy to the well-described cardiac torsion mechanics.17
The problem in the studies of ventricular models is that unlike skeletal muscles, myocardial tissue is locally arranged in a discrete mesh of branching myocytes.18 This entangled structure is prone to hinder or even mislead the interpretation of “tracts” that define the muscular structure of the myocardium. Some researchers argue that the interpretation of such “tracts” depends on the dissection procedure.12
During the last decade, a new imaging modality, diffusion tensor magnetic resonance imaging (DT-MRI), has enabled computational validation of the muscular structure of the heart. This technique provides a discrete measurement of the 3-dimensional arrangement of myocytes19 by the observation of local anisotropic diffusion of water molecules in biological tissues.20 DT-MRI has been established as the reference imaging modality for the measurement of the whole cardiac architecture with acceptable resolution (300μm×300μm×1000μm) compared to the size of myocytes (50-100μm long and 10-20μm thick). Indeed, DT-MRI provides a summary of the microscopic mesh enhancing the preferred pathway of the connected myocytes, which constitutes the concept of myocardial fiber.
In the present study, advanced computer graphics techniques were used to provide an objective and comprehensive description of the myocardial fiber architecture, as previously communicated,21 and we introduce a multiresolution tractographic approach to provide a simplified and comprehensive understanding of the heart architecture.
METHODSDatasets used in this study come from the public database of the Johns Hopkins University.22 These datasets were obtained from 4 normal canine hearts. Each heart was placed in an acrylic container filled with Fomblin, a perfluoropolyether (Ausimon; Thorofare, New Jersey, United States). Fomblin has a low dielectric effect and minimal MRI signal, thereby increasing contrast and eliminating unwanted susceptibility artifacts near the boundaries of the heart. The long axis of the hearts was aligned with the z-axis of the scanner. Images were acquired with a 4-element, knee phased-array coil on a 1.5 T GE CV/I MRI scanner (GE Medical System; Wausheka, Wisconsin, United States) using an enhanced gradient system with 40mT/m maximum gradient amplitude and a 150 T/m/s slew rate. Hearts were placed in the center of the coil and a 3-dimensional fast-spin echo sequence was used to acquire diffusion images with a minimum of 16 noncollinear gradient directions and a maximum b-value of 1500s/mm2. The size of each voxel was about 312.5μm×312.5μm×800μm. Resolution resulting from zero padding in Fourier space allowed us to adapt original image size of 192×192 to 256×256. The final dataset was arranged in about 256×256×108 arrays (depending on the scanned heart) and contains two kinds of data: geometry/scalar data and diffusion tensor data. For diffusion tensor data, each voxel in the array consisted of 3 eigenvalues and 3 eigenvectors. The size of each voxel was about 312.5μm×312.5μm×800μm.
Full-scale tractographies presented in this study have been built with 200 seeds. These seeds were randomly chosen from the entire anatomy, excluding only a very small range of points related to the lowest eigenvalues that are likely to be bad starting points for the reconstruction. The strategy for the seed selection in the reconstructions of lower resolution in the scale-space was to scale these values in proportion to the downscaling magnitude.
Key Points for Ventricular Tractography Reconstruction- •
Data completeness: it is undisputed that the basal ring is crucial to fully understand heart anatomy and function. However, in some publications23–25 the myocardial volume is cut just below the mitral valve to avoid noisy tractography in the auricular cavities. Given that this plane cut discards the basal ring, reconstructions are too incomplete for a reliable interpretation of the cardiac architecture.
- •
DT-MRI vector field orientation: tractography is a technique inherited from the study of fluids, in which the orientation of vector fields stands for fluid stream directions, and thus reconstructions present no ambiguity. However, in the case of anatomical structures the orientation of DT-MRI vector fields does not correspond to any physiological property. For a successful tractography reconstruction, DT-MRI vector fields should be reoriented. The few existing approaches are based on either local properties of the flux or parametric models of the heart. By their local nature, local approaches24 might introduce suboptimal fibers not consistent with the global structure. Although parametric models of the ventricles26,27 provide a good solution to solve fiber orientation, because of their complexity they are usually restricted to the left ventricle. We propose a geometrical organization coherent to gross heart anatomy.
- •
Visualization: comprehensive visualization of fiber tracts should involve a proper assignment of colors providing information about the orientation of the myocardial fibers. Often color maps are defined using a global coordinate system, which might misrepresent the global structure. In order to properly encode the anatomical structure, color maps based on local information should be considered.
- •
Heart architecture interpretation: fully detailed tractographic reconstructions fit perfectly for low-level descriptions, but might fail on a higher level of analysis as a result of their complexity. To obtain more comprehensive descriptions of global myocardial structure, we propose a multiresolution approach applied to the standard tractographic algorithms. This may help to generate simpler visualizations, which in turn may help to better understand the detailed myocardial architecture.
Heart tractography is seen as a reconstruction composed of several streamlines28 (also known as fiber tracks in this field). The main property that clearly defines a streamline is that it is a curve tangential to the vector field at any point of such curve.
In this study, tractographies will be composed of streamlines computed on the vector field of primary eigenvectors at the diffusion-tensor volumes. We computed those streamlines using a fifth-order Runge-Kutta-Fehlbert29 integration method that is able to provide successful results using variable integration steps based on error estimation.
- •
Data completeness: to achieve complete reconstructions of the myocardial anatomy we have considered the whole DT-MRI volumes, including the atrial cavities and the basal ring. Noise on the streamline reconstruction is mainly caused by thin atrial tissue, which introduces significant clutter on the visualization. To minimize this artifact, our streamlining method stops integration of streams with a large Runge-Kutta estimated reconstruction error.
- •
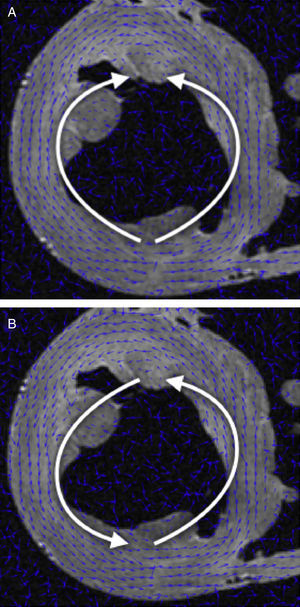
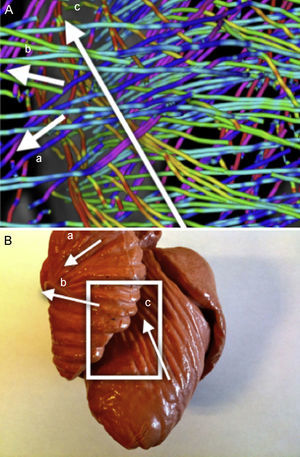
DT-MRI Vector field orientation: tractography is a graphical representation inherited from fluid mechanics, where both direction and orientation of the vector fields are a meaningful part of the represented information. However, on DT-MRI data the vectors can be considered bidirectional because the water diffusion represented by this eigenvector occurs in one dimension but it does so on both possible orientations at the same time. Sometimes the datasets will have a nearly organized structure, but we can also get opposed orientations (Fig. 1A) at some points of the vector field that hinder its reconstruction.
- •
We applied a geometrical reorganization of the vector field using local coordinate systems coherent with ventricular anatomy and fluid mechanics. Ventricular anatomy can be described by a longitudinal axis and angular coordinates with respect to this axis on axial cuts. In order to properly reorient both ventricles, our longitudinal axis was set across the left ventricle, near the septum, ensuring that it never crossed any myocardial wall. To achieve a valid vector field for streamlining, this axis should define a center of rotation for each axial cut. Therefore, at every axial cut of the DT-MRI we reorganize vector orientations in a stream-like fashion (Fig. 1B) around the point where the coordinate axis intersects the same axial cut. This implementation allows fast reorientation, avoiding any smoothing of the vector field.
- •
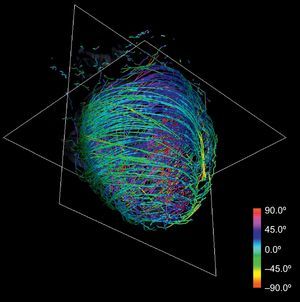
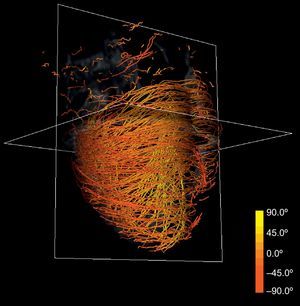
Anatomical-based fiber coloring: the previous reorientation allows coloring techniques based on axial and longitudinal angulations of fibers that may help in the interpretation of the tractographic models. Different color mappings coherent with these directions allow highlighting of different features of the fiber architecture, adding valuable information about existing muscle layers. Color maps tuned to longitudinal angulation convey more valuable information about muscle layers. Figures 2 and 3 show two different views of the longitudinal color map of the reconstructed fibers.
The representation of a fully detailed tractography has been the state-of-the-art methodology to work out the comprehension of the heart. On this task, tractographic models have achieved interesting results but have not been able to define a unique, widely accepted description of myocardial anatomy.
Intuitively, when an observer tries to make a gross analysis in the real world context he can step away a few meters from the object of analysis and get a more contextual view. We will extrapolate this everyday behavior to our problem.
In order to resolve this in a computer graphic representation it is common to use multiresolution models, which attempt to build different models of the same data with different levels of detail but without a loss of fidelity. Usually applied to texture mapping, this technique is known as mipmapping,30 based on the well-known pyramid representation.31 This technique applies a Gaussian filtering and later an exponential reduction via a subsampling of the full-scale texture. Reduced textures are “summaries” of the original texture and are used to represent this texture at different scales. These “summaries” are statistically complete so that the Gaussian smoothing keeps the contextual information before applying downsampling. The use of these downscaled images is also common in other fields such as computer vision, where this operation can be seen as a computation on the scale space.
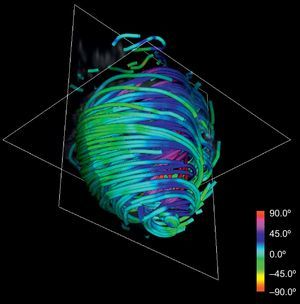
This technique can be applied to the DT-MRI dataset to simplify its complexity. By downscaling two orders of magnitude of the original datasets and applying our streamlining, we get the simplified tractography shown in Figure 4. In comparison with the full-scale tractography shown in Figure 2, the simplified one keeps the main geometric features of fibers. Therefore, it allows easier identification of global morphological tendencies.
RESULTSOur simplified tractographic reconstruction method (Fig. 4) keeps the main geometric features of fibers, allowing an easier identification of overall trends. In turn, these trends show a manifest continuous helical structure of the ventricular myocardium. We sought to compare the results of the tractography with the HVMB anatomy described by Torrent-Guasp et al.15
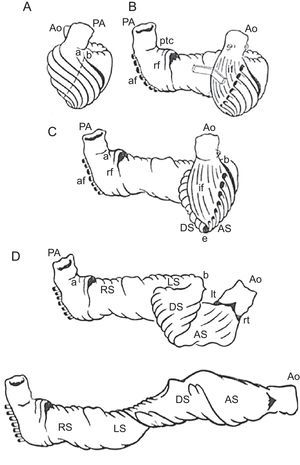
The HVMB model describes a longitudinal arrangement of ventricular myocardial fibers forming a unique functional muscleband (Fig. 5) starting at the pulmonary artery and finishing at the aorta. This muscle wraps the left ventricle and part of the right ventricle (right and left segments), connecting to a helicoidal structure starting at the basal ring going inside the left ventricle towards the apex and returning to connect with the aorta (descending and ascending segments), with this turn wrapping the entire anatomy of the heart.
Ventricular myocardial band. Schematic presentation of the ventricular myocardial band dissection. af, aberrant fibers; Ao, aorta; AS ascending segment; DS, descending segment; if, intraseptal fibers; LS, left segment; lt, left trigone; PA, pulmonary artery; ptc, pulmonary-tricuspid cord; rf, right septal fibers; RS, right segment; rt, right trigone.
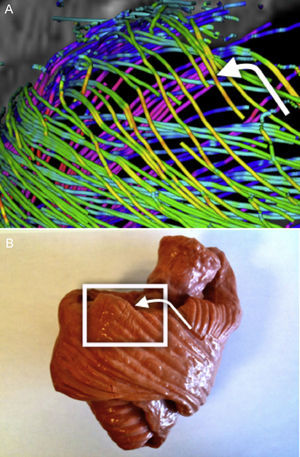
To compare tractographic results with the band model, step-by-step tractographic reconstructions were compared with the myocardial fiber tracts depicted in the Torrent-Guasp rubber-silicone mould of the HVMB32 (Figs. 6–9).
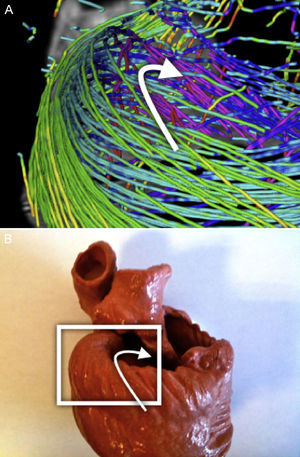
A clear pattern is observed where the reconstructed tracts on the epicardium are oriented towards the basal ring. These tracts loop at the basal ring toward the endocardium, describing what looks like a simple folding (Fig. 6). As we track through lower streamlines, the lines are organized more horizontally but preserving a slight slope. We can see that these lines describe trajectories that wrap around the left ventricle and connect to further folds at the basal ring (Fig. 7).
Left SegmentThe previous pattern is reproduced along the left segment. At the end of this segment we can observe that the mentioned folding ends at the point where the streams get into the endocardium (Fig. 7).
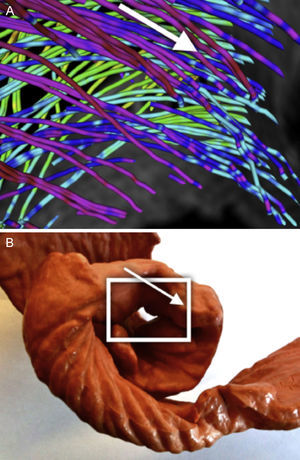
Descending SegmentFrom an anterior view (Fig. 8) we can clearly distinguish a spiral-descending organization of the endocardium population of streams across the septum. This structure continues to the apex and most of these streams continue on the right segment. Behind this endocardial structure an ascending structure is visible that we will analyze in the following section from another visualization point of view.
Ascending SegmentThe analysis of this segment is more complex due to the cluttered view of several crossings of myocyte populations. With fewer streamlines than on the previous captures, Figure 9 shows 3 populations where in this area streams coming from the apex start a noticeable ascent (fading from green to red coloration of the streams, denoting an increased slope) below the two other populations that are the beginning of the right segment at its connection with the pulmonary artery.
Simplified TractographyAlthough our simplified models provide easier interpretation of global trends, they are still too complex for summarizing complex structures such as the Torrent-Guasp HVMB.
To simplify the backbone myocardial fiber spatial orientation, we explored the geometry of the heart by looking for long paths that can represent connected regions on the DT-MRI tractography. The goal of this procedure was to provide a comprehensive reconstruction that allows interpretation at first sight by any possible observer.
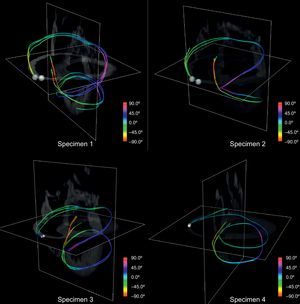
By manually picking seeds at the basal level we obtained continuous paths connecting both ventricles and wrapping the whole myocardium. Figure 10 shows 4 tracts of simplified models reconstructed from manually picked seeds located at basal level near the pulmonary artery. We observed that the tracts define a sample-wide coherent helical structure for all canine samples.
The use of visualizations with single tracts changes the way in which this structure can be viewed. We compared such tracts to the proposed HVMB (Fig. 11). There is a clear similarity between the HVMB schematic model (Fig. 11, left) and reconstructed paths (Fig. 11, right). In both models the main segments (labeled from A to G) of the helical architecture are clearly identified.
Comparison between helical ventricular myocardial band scheme and a simplified ventricular tractography. The Torrent-Guasp helical ventricular myocardial band model (left) compared to a tract reconstructed (right) from a single manually picked seed on the diffusion tensor magnetic resonance imaging volume with landmarks for comparison with the model.
The present paper provides an objective interpretation of the myocardial architecture based on automated DT-MRI descriptions. Results show an unequivocal ventricular fiber connectivity describing a continuous muscular structure consisting of the two ventricles arranged in a double helical orientation. This supports the Torrent-Guasp description of the HVMB. These results are shown by unique, automatically-generated tracts that describe this connectivity along the whole myocytal mesh starting at the pulmonary artery and finishing at the aorta.
The DT-MRI technique provides trustworthy and detailed information of myocardial tissue. However, interpretation of its outcome for heart architecture validation is indirect. Existing techniques reconstruct full heart anatomy using visual cues. Since tractography was first proposed and used for heart structure study,23,33 it has been the most common technique to recover information from DT-MRI. Other techniques also have been explored, such as those in the work of Frindel et al.25 based on the optimization of graph models that promise future developments.
There are many factors that should be taken into account in order to obtain widely acceptable reconstructions and interpretations. It follows that most of the existing approaches23–26,34,35 do not provide enough evidence widely accepted by the whole scientific community to either support or invalidate any particular architectural model. The only agreement is the existence of a layered structure of the myocardium through tractographic representations and visualization improvements in color coding. Among these efforts, we would highlight the work of Helm et al.26 since, due to its level of detail, it has been widely discussed in the literature hinting at opposite readings. Such disagreement is a direct consequence of a partial reconstruction of the heart fiber anatomy.
In order to settle this disagreement we used all the DT-MRI data without segmentation to avoid instrumentalization of the study, and demonstrated that it is possible to reconstruct the whole myocardium including some complex structures such as the basal loop, unfortunately hidden or misinterpreted by other studies. It was also necessary to define a method that ensures a correct use of streamlining techniques to the particularities of the DT-MRI vector fields.
Validation of the correctness of local structures is not enough to extend the interpretation to a global point of view. To deal with higher-level interpretations of the architectural organization of the heart we also looked for higher-level representations that can ease its interpretation and validation. We have contributed a multiresolution method for tractography using downsampling of the DT-MRI volumes to show overall features of the heart structure. This work also includes coloring techniques applied to our solution to ease the reading of the tractographic 3-dimensional models.
For studies requiring Q-ball analysis it is mandatory to use not less than 60 directions per voxel. However, diffusion tensor imaging (DTI) tensors only provide an average description of water diffusion and thus a large number of diffusion directions do not significantly improve their quality. It follows that existing DTI cardiac studies (like the widely used Johns Hopkins University data set36) for DTI analysis usually restrict values between 12 and 32 directions37 for the sake of a good compromise between acquisition time and quality. Furthermore, a recent study reports that the DTI primary eigenvector is invariant under a large variation of acquisition device parameters and, in particular, to a low number of diffusion directions.38 Our own research suggests that heart preparation and volume spatial resolution are, indeed, one of the most influencing conditions on DTI quality. Acquisition field-of-view should be carefully adjusted to fit just the myocardial volume, which should be in suspension inside a recipient in order to avoid distortions in diffusion near myocardial boundaries.
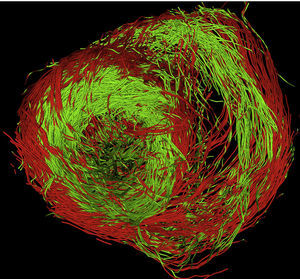
We are currently acquiring our dataset from pig hearts using a 3T Philips device with 32 gradients, a volume resolution of 1.38μm×1.38μm×1.5μm (144 voxels×144 voxels×60 voxels) covering a heart short-axis region of interest of 70 pixels×70 pixels. Figure 12 shows a full-resolution tractographic reconstruction of muscle fibers obtained using our software. The coloring indicates the sign of the fiber z-component (red for positive and green for negative) and, thus, its orientation. The transition fiber loop from epicardium to endocardium is clearly seen in the left lateral segment of the left ventricular base. The conclusions in this paper show the high quality of DT-MRI heart study.
CONCLUSIONSThe objective analysis of myocardial architecture by an automated method including the entire myocardium and using several 3-dimensional levels of complexity reveals a continuous helical myocardial fiber arrangement of both right and left ventricles, thus supporting the anatomical studies performed by F. Torrent-Guasp.
FUNDINGThis work was supported by the Spanish TIN2009-13618 and TIN2012-33116.
CONFLICTS OF INTERESTNone declared.
We want to acknowledge Drs. Patrick A. Helm and Raimond L. Winslow at the Center for Cardiovascular Bioinformatics and Modeling and Dr. Elliot McVeigh at the National Institute of Health for provision of DT-MRI datasets.